Viral taxonomy and phylogeny I
Overview
Teaching: 60 min
Exercises: 120 minObjectives
Understand the differences between taxonomic approaches for viral and cellular organisms
Explain the challenges with viral taxonomy, and how they may be overcome
How are viruses taxonomically classified?
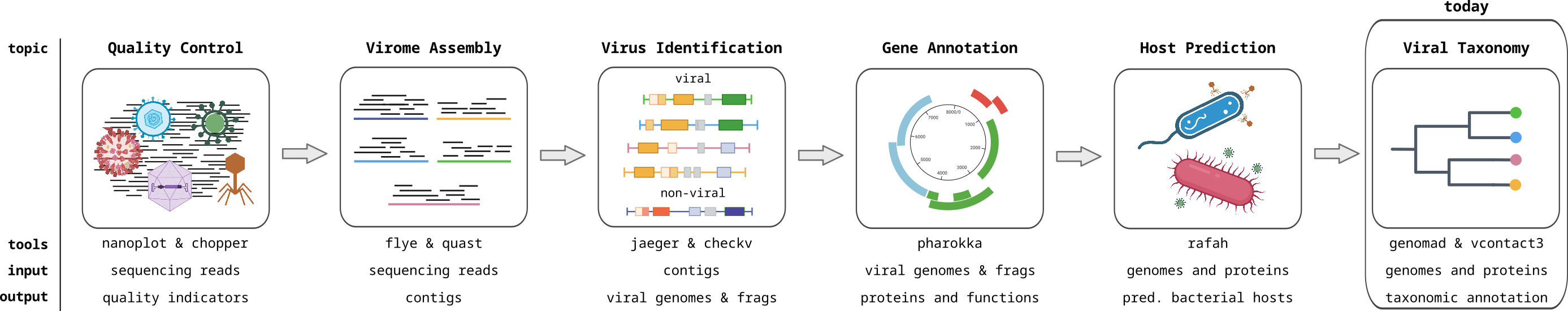
There is no single method to classify the taxonomy of viruses. Many experts from the global virology community have done their part by classifying viruses according to their specific knowledge. This has generated a patchwork of methods that capture the features of different viral lineages and generate meaningful taxa that are in agreement with biology. With the advancement of viromics and the discovery of viruses by their genome sequences only, new methods are necessary to classify viruses based on their sequences, similar to approaches for cellular organisms.
A widely accepted approach for sequence-based taxonomic classification is by selecting a marker gene that is shared by all organisms, creating a multiple sequence alignment and phylogenetic tree, and identifying taxa as characteristic branches or lineages in the tree. This approach can be applied to all cellular organisms including bacteria, archaea, and eukaryotes, particularly using with ribosomal genes such as 16S small subunit in prokaryotes or the 18S in eukaryotes. As no marker gene is universally conserved in all viruses, this approach is only possible in subgroups of viruses. The lack of a universal genomic feature is thought to reflect their multiple evolutionary origins.
Bioinformaticians have developed many methods to circumvent the lack of a universal gene, for example, by cluster viral sequences. However, so far these methods have not been widely adopted for official ICTV virus taxonomy.
One popular method in the bacteriophage field is the gene-sharing network. This involves creating a network where nodes are viral genomes and edges represent shared gene families between these genomes. Tight clusters in this gene-sharing network represent groups of viral genomes that share many genes, and those could be interpreted as taxonomic groups. Networks can be built with different gene-sharing cutoffs, corresponding to different taxonomic ranks, where closely related viruses (species, genera) share more genes than distant ones (families). Tools like VICTOR and vConTACT3 work this way.
As more and more viruses are sequenced and we get a better view of the virosphere, marker genes are making a comeback. Although a universal marker gene shared by all viruses does not exist, marker genes are certainly shared by viruses of lower-ranking taxa (species, genera, families, orders, and in some cases above). Detecting the marker gene is evidence that a virus belongs to a given taxon, and phylogenetic trees can help resolve the lower-level taxonomy, just like ribosomal marker gene trees for cellular organisms. Tools like vClassifier and geNomad use marker gene approaches.
In the practical section of this day, we will be using marker gene methods and building a gene sharing network to classify the viruses identified in our metaviromes. Examples of widely used marker genes are for phages include the terminase large subunit (terL), major capsid protein (mcp) and DNA polymerase B (DnaB).
Exercise - An overview of viral taxonomy
- Check out the ICTV website: https://ictv.global/.
Discuss with your fellow students and write down your answers:
- What is ICTV? (not just a TV channel featuring polar bears)
- How many taxonomic ranks are available to classify the virosphere?
- What is a realm?
- How many viral genera are there?
- Pick one marker gene and describe how it functions in the virus-host interaction, which taxonomic rank it can/cannot identify.
Read the following sections from the review “Global Organization and Proposed Megataxonomy of the Virus World” by Koonin et al.:
- The Baltimore classes of viruses, virus hallmark genes, and major evolutionary trends in the virus world
- Evolutionary links among viruses within and across the Baltimore classes, section: Double-Stranded DNA Viruses
Discuss with your fellow students and write down your answers:
- What are the Baltimore Classes, and what are they based on?
- What triggered the change in viral taxonomy?
- What are the pros and cons of a “genomic taxonomy”?
Key Points
Viruses have multiple origins, so there is no universal marker gene
Viral taxonomy is based on many different methods
Gene-sharing networks and marker genes are popular methods for bacteriophage taxonomy