Inspecting the MSAs
Overview
Teaching: 10 min
Exercises: 30 minQuestions
Quality of the multiple sequence alignment
If the MSAs we base our search on are of poor quality we cannot expect to find matches when querying the PHROG database.
MSA quality
- Think of what factors will influence the quality of our MSAs
For a small set of proteins we can manually inspect the MSAs, however, for thousands of proteins this will take too long. Hence I used MstatX to automatically assess the quality of the MSAs.
Among one of the factors that might influence the quality of our MSAs is our clustering using mmseqs2 - the input for mafft. Specfically the parameters we chose, like the sensitivity (-s), and also the default values for identity (--min-seq-id) and coverage(-c). Ideally we would try different values and carefully read the manual on how to choose the most suitable parameters (e.g. here for the coverage). For now I ran MstatX on all the MSAs from the other step. You can see the distribution here:
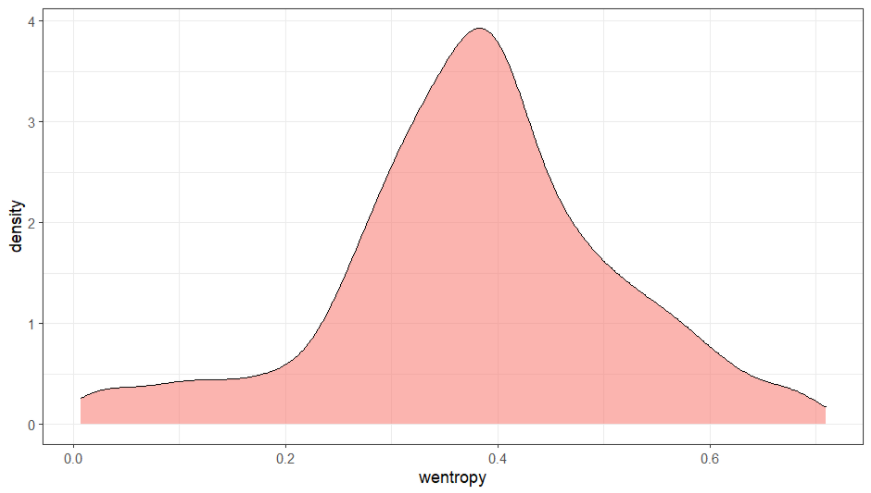
You can get the MSA for the highest entropy MSA here and the lowest entropy MSA here. Install Jalview from here and visualize the alignments.
Entropy values
- In Jalview, can you explain the high and low entropy values?
- Do you think high, middle or low entropy would be the best to do a model search?
Key Points