Getting to know your files
Overview
Teaching: 120 min
Exercises: 0 minObjectives
Interpret different file formats used in bioinformatics
Utilize bash commands to look into sequence data
Get a basic understanding of what sequence data looks like
Work with sequence data in Python or R to create basic plots
Navigating your filesystem space
First, let’s copy the sequence data to your own home folders.
Exercise - get your sequence data
The data is in the folder
/work/groups/VEO/shared_data/Viromics2024Workspace/data/sequences/viral_metagenome# make a ./data/sequences directory if it doesn't already exist IN YOUR OWN HOME DIRECTORY mkdir -p viromics/data/sequences # go into your sequences directory cd viromics/data/sequences # copy over the 3 barcode files you need cp /work/groups/VEO/shared_data/Viromics2024Workspace/data/sequences/viral_metagenome/full_barcode*.fastq.gz ./
Exercise - check out your files
- Take a look at the file structure and files. We have given you a subset of viromics data
./data/sequences. Some helpful commands might be: Hint: for gzipped files - you need to zcat them first. e.g.zcat file.fastq.gz | head -10# list ls ll # path to working directory pwd # print first 10 lines zcat file.fastq.gz | head -10 # print last 10 lines zcat file.fastq.gz | tail -10 # open file on terminal (press 'q' to exit less) less more # count number of lines wc -l file.txt # search for "@" inside file grep "@" file.fastq # or gzipped files zgrep "@" file.fastq.gz | head -10 # count number of sequence headers in a fasta file grep -c ">" file.fasta zgrep -c ">" file.fasta.gz
See more useful commands and one-liners here
For creating new directories - please ensure to name your directories with a number and a meaningful header. An example is here
Understanding bioinformatics file formats
It is always a good idea to check the contents of your data files and make regular “sanity checks” to see if you understand everything. The first video covers different file formats commonly used in bioinformatics. Namely, FASTA, FASTQ, BAM, SAM, VCF, BCF, GFF, GTF and BED files (9 minutes).
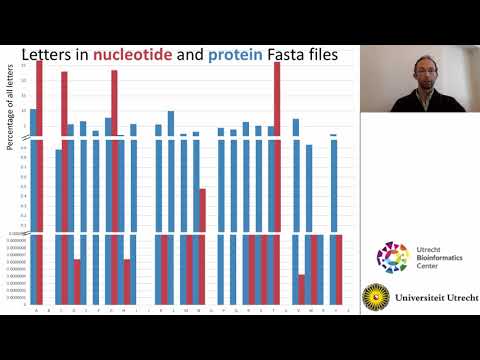
The second video covers different types of sequence files, including fasta and fastq, phred scores, files containing metadata (tsv files) and compressed files (11 minutes).
Exercise
- What information is contained in sequencing files?
a) Choose 3 file formats and describe them- Choose the barcode62.fastq.gz file in your data/sequences/ folder
a) How many lines does this file have?
b) How many sequences are present in the file?- Print the first 5 lines and the last 5 lines in the terminal.
Hint: for gzipped files - you need to zcat them first. e.g.zcat file.fastq.gz | head -10
You don’t need to print this output into your lab-book
zcat barcode62.fastq.gz | cut -b -100 | head -10- to print the first 100 characters of the first 10 lines
a) What differences do you observe between these lines?
Key Points
Sequence data and information about genes and genomes can come in many different formats
The most common file formats are fasta (nucl. and amino acid), fastq, sam and bam, genbank, gff and tsv files