Here you can find information about the advanced module “Viromics Bioinformatics” of the Masters program in Microbiology of the Friedrich-Schiller University of Jena.
It consists of 60 hours dry lab work to be completed in two weeks.
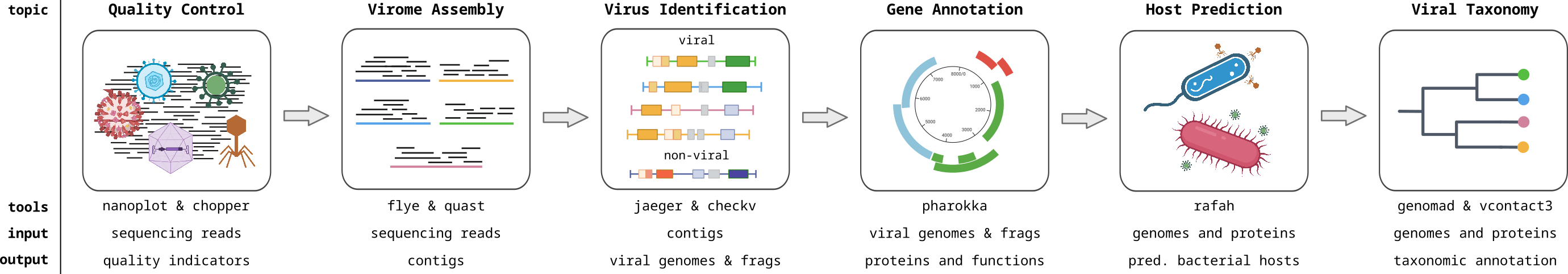
You will learn how to analyse bacteriophage sequences with computational approaches, covering the following topics:
- Sequencing quality control
- Assembly of long-read sequences
- Identifying viral contigs
- Open reading frame (ORF) predictions
- Gene annotation
- Host prediction
- Viral taxonomy and phylogeny
- Visualizations
At the end of the module, you will have the opportunity to present your work to your classmates and the teachers.
If you do not have a solid background in linux/bash, please follow this self-study material before the start of the module.
At the top of the webpage, under “Extras”, you will find notes on: