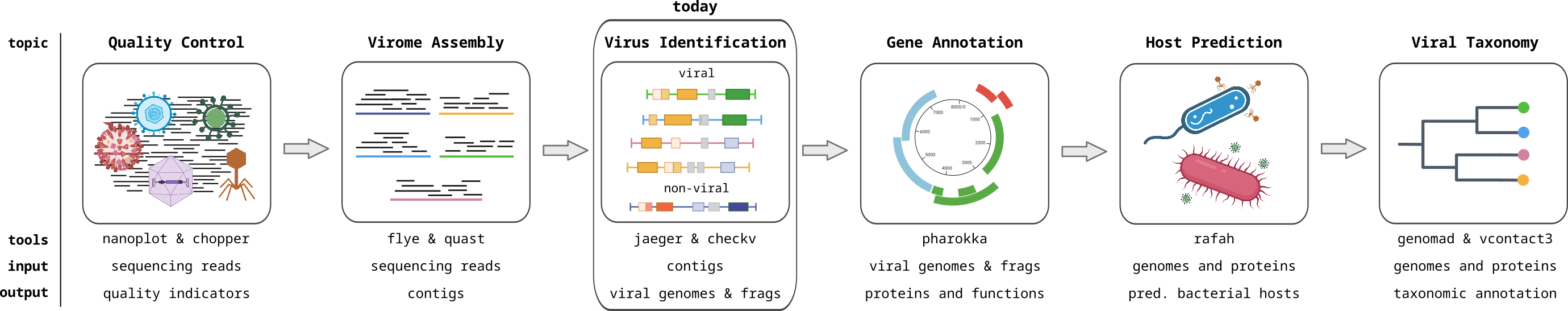
Identifying Viral Contigs I
Overview
Teaching: 0 min
Exercises: 180 minObjectives
Understand the concept and structure of benchmarking virus identification tools
Evaluate the benchmarking results presented in the paper by Wu et al
Choose a virus identification tool
Comparing Virus Identification Tools
In this theoretical part, you will dive into a very important part of microbiome data analysis: how to choose a good tool. Several tools have been developed over the past years for identifying viral sequences. Each tool measures different aspects of the sequence, applies different algorithms to extract and compare the information, and uses different reference databases to distinguish viral from non-viral sequences.
To choose the best one for your project, you should consider the overall performance of the tools (e.g. sensitivity and specificity, but also ask whether the purpose of the tool aligns with your research goal. If a recent tool does the same task as a previously published one, the authors should compare (or benchmark) them in their publication. But benchmarks can be biased, so ideally, bioinformatic tools are benchmarked by independent scientists.
Today, we will study the paper: “Benchmarking bioinformatic virus identification tools using real‐world metagenomic data across biomes” (Wu et al., 2024). Considering the allocated time for this activity, you will probably not be able to read the entire paper. To answer the proposed questions, please focus on Figures 1, 3, and 6 (and associated Methods and Results paragraphs, search online when necessary).
Questions
The authors investigated nine tools, which can be divided into three categories. What categories? Describe the basic approach of each category of tools.
What is meant by the sets of True Positives, False Negatives, False Positives, and True Negatives in Figure 1? Explain what each of these sets can tell us in the benchmark, and explain how they were generated.
In Figure 3 A/B/C, what indicates a good performance?
In Figure 3 D/E/F, the terms TPR, FPR, and AUC/ROC are used for evaluation. What are those terms? What is the main takeaway from these figures?
Overall, which types of tools perform better? Why do you think that is?
Explain what is shown in Figure 6. Do you believe it? What value does this analysis add to the benchmarking?
Considering all you have learned from the paper, is there an ideal tool to identify viral sequences? Why (not)?
For the project we are developing in this module, which tool(s) would you use? Why?
Key Points
Different approaches can be used in a virus identification tool, such as reference-based and machine learning
Benchmark is a comparison between tools and should offer concrete evidence of performance, such as number of TP, FN, FP and TN
When choosing a tool, you should consider the priorities of the project. E. g. you need to identify as many viruses as possible and FP are not critical, so a tool with high recall is ideal