Gene Calling and Functional Annotation I
Overview
Teaching: 120 min
Exercises: 60 minObjectives
Understand what is gene calling and functional annotation
Investigate the features of phages and how it impacts ORF annotation
Understand what information functional annotation can provide
Gene Finding
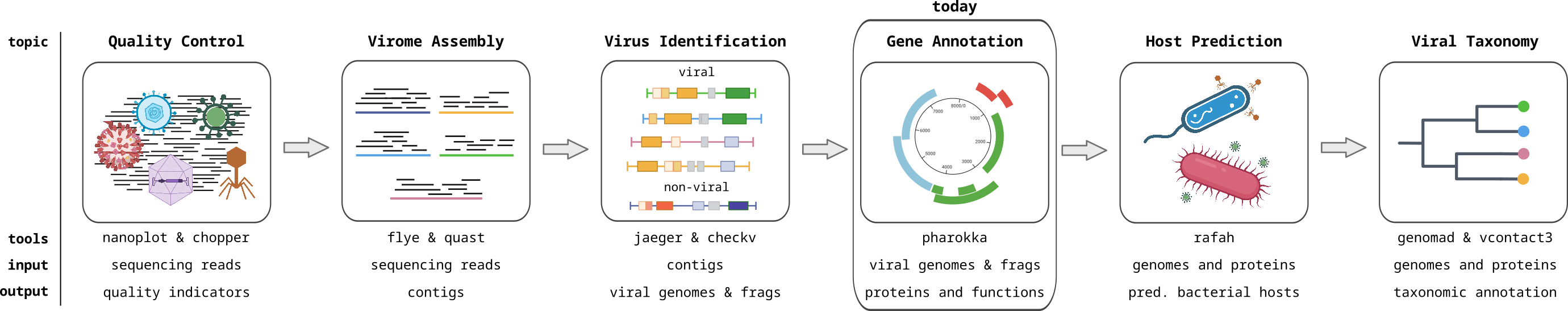
Once we have an assembled genome sequence (or even a contig that represents a fragment of a genome), we want to know what kind of organism the sequence belongs to (taxonomic annotation), what genes it encodes (gene annotation or gene calling), and the functions of those genes (functional annotation). Ideally, gene calling uses a good gene model that is tailored to the organism of study, but to determine the organism we need to know its genes. So it can be an iterative process. In our case, there are pretty good standard gene models available for bacteria and for phages, which we will use.
Phage genomes have specific features that are important to keep in mind. The following lecture by Dr. Robert Edwards explains these, and how they were taken into account when developing a specialized phage gene caller Phanotate.
- Click on the image to see the lecture video “Phage Genomics” by Dr. Rob Edwards (29 minutes):
The video below by Dr Evelien Adriaenssens explains more about phage genome structure and functional annotation
- Click on the image to see the lecture video “Basics of phage genome annotation & classification: getting started” by Dr Evelien Adriaenssens (68 minutes, watch from 12:02 to 30:45):
Exercise
Watch the video lectures for gene finding and functional annotation to answer the questions below:
- What is genome annotation?
- What are ORFs and why are they important in annotating genes?
- Explain the Phanotate algorithm.
- Name at least 3 features of phages should be considered when developing a phage gene annotation tool
Additional Resources
The links of Prodigal and Phanotate might also useful for answering the questions.
As additional material, the video by Dr. Katelyn McNair contains explanations of the algorithm of Phanotate. The topic is similar to the video by Dr. Robert Edwards, but more detailed about the algorithm
Functional Annotation
Once you have ORFs for your phage genomes, you might be curious to what genes they represent. Annotating genes on a genome is called functional annotation and gives insights into the phage lifestyle (lytic or lysogenic), taxonomy, hosts interactions and much more. Therefore, this is probably one of the most biologically informative and interesting steps of the viromics pipeline.
To annotate the phage genomes we will use a tool called Pharokka. Pharokka uses phannotate for the gene calling.
Exercise
The pharokka paper.
Please read the paper for pharokka - from sections 1 - 2.4 to answer the following questions:
- What genomics features are taken into account in the pharokka algorithm?
- How does pharokka assign functional annotations to ORFs using PHROGs?
- What is an HMM?
Additional Resources
The PHROG paper and the PHROGs database might also be useful for you
A caveat to pharokka
As pharokka uses PHROGs to annotate viral genomes, it is limited in the types of auxiliary metabolic genes it can assign. This limitation is set by which genes were previously classified as AMGs and therefore have a PHROG. De novo assignment of AMGs is not possible. AMGs therefore might be better assigned by using a bacterial or a combination viral+bacterial functional annotation tool like DRAM
Key Points
Genome annotation gives meaning to genomic sequences
ORFs can be predicted from start and stop codons in the genomic sequences
Phages have different genomic features than prokaryotes, which influences the design of algorithms
Tools like Phanotate are very useful to process a large amount of contigs. However, no tool is perfect, so a critical interpretation of the results is important
Functional annotation of viral genomes can give clues about viral lifestyle and host interactions